This blog is dedicated to those who carry the J2 "Y" DNA Haplogroup, with a focus on J2a4h2, also known as J-L25, with further changes to F3133, and FGC9962. Our "Y" Chromosome is inherited from father to son. Our paternal ancestors will also have the same signature. "J2 originated in northern Mesopotamia, and spread westward to Anatolia and southern Europe, and eastward to Persia and India.
Y Chromosome Information - passed from father to son. It is not clear how our ancestors got to Switzerland, although with more people getting the test and more research a detailed path may yet be found.
This blog tracks the DNA Y chromosome for the Hullinger / Hollinger / Holliger / Holiger clan. As we find new information we will add it to the blog while keeping this summary updated.
Our oldest know paternal ancestor is Henri Holiger of Boniswyl, Aargau, Switzerland, born in 1425. We have visited this lovely area and found relatives. The community is very attractive with a small castle located at the foot of a lake. A fast flowing stream forms a moat around the castle.
Our paternal Swiss ancestors immigrated to the United States in 1736 and settled in Pennsylvania. Their descendants gradually moved west with the frontier. Our branch of the family homesteaded in Vivian, South Dakota in the early 1900's.
We conducted genetic testing to learn about our ancient history. Craig Hullinger was tested for the "Y" chromosome which is passed from father to son with very little change. Clif Hullinger was tested for mtDNA, which is passed from mother to child. The results of Clif Hullinger's test is at harlandna.blogspot.com
Our Male "Y" Haplogroup is J2A4H2. This is not a widespread Haplogroup in Switzerland. It originated in the middle east.The maps below show the origination and migration path of men with the "J" Haplogroup.
"Y-DNA haplogroup J2 lineages originated in the area known as the Fertile Crescent. The main spread of J2 into the Mediterranean area is thought to have coincided with the expansion of agricultural peoples during the Neolithic period. "
"J2 is related to the Ancient Etruscans, (Minoan) Greeks, southern Anatolians, Phoenicians, Assyrians and Babylonians. In Europe, J2 reaches its highest frequency in Greece (especially in Crete, Peloponese and Thrace), southern and central Italy, southern France, and southern Spain. The ancient Greeks and Phoenicians were the main driving forces behind the spread J2 around the western and southern Mediterranean."
So our Swiss ancestor came from the middle east. Other Hollinger and Hullinger's have also conducted testing. As more people conduct tests we will learn more.
The test showed that our Swiss Hullinger genealogy was accurate. Our "Y" chromosome is closely related to other Hullinger / Hollinger men who who also took the DNA test. The table below shows twelve men with the Hollinger name who took the DNA test.
Eight are closely related (My test is the one indicating Henri Holiger). Four of the eight list their ancestral country of origin as Switzerland, with two unknown and one each from Austria and Germany. The other eight men are not closely related to us and perhaps acquired their last name through adoption or developed independently in a different locale, or they are related to Hollinger on the female line.
It thus seems likely that our ancestor immigrated into Switzerland and then acquired our last name. Our Hollinger (Hullinger) Name, according to Genealogy Family Education. http://genealogy.familyeducation.com/
South German and Jewish (Ashkenazic): habitational name for someone from places called Holling or Hollingen.
The test also shows the number of Swiss matches against the total number of Swiss and European "Y" Haplogroups tested. This result is quite low - only 2 Swiss cousins out of 1,618 Swiss tested. It is also rare in the rest of northern Europe.This indicates that our J2 haplogroup is a fairly recent and rare haplogroup in northern Europe.
Other DNA tests listed later in this blog show the total number of men who tested J2A4H2. Many of them are from the middle east.
We also found another interesting item. There were a large number of European Jews named Hollinger who were killed in the Holocaust. We don't know the connection. The Jewish Hollingers could have acquired their name independently of ours or they could be closely related. We will eventually find out - there are a number of Hollingers in Israel. As they get tested for their Y chromosome we will find out if we are closely related. If so our paternal ancestor was most likely Jewish. If not then the Roman soldier or slave solution becomes more likely.
Genetic testing is relatively new. We will likely find out much more about our ancient history as more people get tested and as we learn more about genetics.
Born Died First Last Name Spouse Birth Place / Comments
1425 1504 Henri Holiger Boniswyl, Aargu, SWZ
1446 Heini Holiger Boniswil (Holvil) Switzerland
1472 Hans Holiger 1504 Junghans Holiger
m Margaretha Rebmeyer
1548 1600 Heini Holiger m Barbara Mayer Boniswyl,Aargu, SWZ Burial: Seengen
1591 1643 Heini Holiger m Anna Huber
Aargu, Boniswyl, SWZ
1627 1689 Rudolph Holliger m Anna Hummel
1661 Jacob Holliger m Elisabeth Burger
1701 1779 Hans Jacob Hollinger m Anna Elisabetha Esterli
Immigrated to US 1736
1734 1802 Christian Hollinger m Eva Dorothea Feltz
Born Germany, Captain American Revolution
1757 1839 Daniel Hullinger m Ann Schockey
Lancaster Co, PA, 1st Lt American Revolution
1788 1856 Daniel Jnr Hullinger m Comfort Conway Staunton Trenton, OH
1833 1909 Daniel J Hullinger m Mary Kirk Ohio emigrated from Ohio to south central Iowa by wagon train in 1864
1870 1956 Eli Hullinger m Mary Elizabeth Siddons Leon IA
1893 1970 John Franklin Hullinger m Pearl Josephine Harlan
Leon, Iowa US Army, WW I
1920 Clifford Harlan Hullinger m Louise Liffengren
Vivian, SD 1st Lieutenant, US Army, WW II
1947 Craig Harlan Hullinger m Elizabeth S. Ruyle
Brookings, SD Colonel, US Marine Corps Reserve, Vietnam
1980 Bret Schaller Hullinger Harvey, IL
There is a lot of additional information in this blog and on the
http://hullingerheritage.blogspot.com/
Our oldest know paternal ancestor is Henri Holiger of Boniswyl, Aargau, Switzerland, born in 1425. We have visited this lovely area and found relatives. The community is very attractive with a small castle located at the foot of a lake. A fast flowing stream forms a moat around the castle.
Our paternal Swiss ancestors immigrated to the United States in 1736 and settled in Pennsylvania. Their descendants gradually moved west with the frontier. Our branch of the family homesteaded in Vivian, South Dakota in the early 1900's.
We conducted genetic testing to learn about our ancient history. Craig Hullinger was tested for the "Y" chromosome which is passed from father to son with very little change. Clif Hullinger was tested for mtDNA, which is passed from mother to child. The results of Clif Hullinger's test is at harlandna.blogspot.com
Our Male "Y" Haplogroup is J2A4H2. This is not a widespread Haplogroup in Switzerland. It originated in the middle east.The maps below show the origination and migration path of men with the "J" Haplogroup.
"Y-DNA haplogroup J2 lineages originated in the area known as the Fertile Crescent. The main spread of J2 into the Mediterranean area is thought to have coincided with the expansion of agricultural peoples during the Neolithic period. "
"J2 is related to the Ancient Etruscans, (Minoan) Greeks, southern Anatolians, Phoenicians, Assyrians and Babylonians. In Europe, J2 reaches its highest frequency in Greece (especially in Crete, Peloponese and Thrace), southern and central Italy, southern France, and southern Spain. The ancient Greeks and Phoenicians were the main driving forces behind the spread J2 around the western and southern Mediterranean."
So our Swiss ancestor came from the middle east. Other Hollinger and Hullinger's have also conducted testing. As more people conduct tests we will learn more.
The test showed that our Swiss Hullinger genealogy was accurate. Our "Y" chromosome is closely related to other Hullinger / Hollinger men who who also took the DNA test. The table below shows twelve men with the Hollinger name who took the DNA test.
Eight are closely related (My test is the one indicating Henri Holiger). Four of the eight list their ancestral country of origin as Switzerland, with two unknown and one each from Austria and Germany. The other eight men are not closely related to us and perhaps acquired their last name through adoption or developed independently in a different locale, or they are related to Hollinger on the female line.
South German and Jewish (Ashkenazic): habitational name for someone from places called Holling or Hollingen.
The test also shows the number of Swiss matches against the total number of Swiss and European "Y" Haplogroups tested. This result is quite low - only 2 Swiss cousins out of 1,618 Swiss tested. It is also rare in the rest of northern Europe.This indicates that our J2 haplogroup is a fairly recent and rare haplogroup in northern Europe.
Other DNA tests listed later in this blog show the total number of men who tested J2A4H2. Many of them are from the middle east.
We also found another interesting item. There were a large number of European Jews named Hollinger who were killed in the Holocaust. We don't know the connection. The Jewish Hollingers could have acquired their name independently of ours or they could be closely related. We will eventually find out - there are a number of Hollingers in Israel. As they get tested for their Y chromosome we will find out if we are closely related. If so our paternal ancestor was most likely Jewish. If not then the Roman soldier or slave solution becomes more likely.
Genetic testing is relatively new. We will likely find out much more about our ancient history as more people get tested and as we learn more about genetics.
Our Paternal Line of Descent
Hullinger / Hollinger / Holliger / Holiger
Born Died First Last Name Spouse Birth Place / Comments
1425 1504 Henri Holiger Boniswyl, Aargu, SWZ
1446 Heini Holiger Boniswil (Holvil) Switzerland
1472 Hans Holiger 1504 Junghans Holiger
m Margaretha Rebmeyer
1548 1600 Heini Holiger m Barbara Mayer Boniswyl,Aargu, SWZ Burial: Seengen
1591 1643 Heini Holiger m Anna Huber
Aargu, Boniswyl, SWZ
1627 1689 Rudolph Holliger m Anna Hummel
1661 Jacob Holliger m Elisabeth Burger
1701 1779 Hans Jacob Hollinger m Anna Elisabetha Esterli
Immigrated to US 1736
1734 1802 Christian Hollinger m Eva Dorothea Feltz
Born Germany, Captain American Revolution
1757 1839 Daniel Hullinger m Ann Schockey
Lancaster Co, PA, 1st Lt American Revolution
1788 1856 Daniel Jnr Hullinger m Comfort Conway Staunton Trenton, OH
1833 1909 Daniel J Hullinger m Mary Kirk Ohio emigrated from Ohio to south central Iowa by wagon train in 1864
1870 1956 Eli Hullinger m Mary Elizabeth Siddons Leon IA
1893 1970 John Franklin Hullinger m Pearl Josephine Harlan
Leon, Iowa US Army, WW I
1920 Clifford Harlan Hullinger m Louise Liffengren
Vivian, SD 1st Lieutenant, US Army, WW II
1947 Craig Harlan Hullinger m Elizabeth S. Ruyle
Brookings, SD Colonel, US Marine Corps Reserve, Vietnam
1980 Bret Schaller Hullinger Harvey, IL
There is a lot of additional information in this blog and on the
http://hullingerheritage.blogspot.com/
Hullinger "Y" DNA Results
| User ID | Last Name | Origin | 3 9 3 | 3 9 0 | 1 9 | 3 9 1 | 3 8 5 a | 3 8 5 b | 4 2 6 | 3 8 8 | 4 3 9 | 3 8 9 | 1 | 3 9 2 | 3 8 9 | 2 | 4 5 8 | 4 5 9 a | 4 5 9 b | 4 5 5 | 4 5 4 | 4 4 7 | 4 3 7 | 4 4 8 | 4 4 9 | 4 6 4 a | 4 6 4 b | 4 6 4 c | 4 6 4 d | 4 6 0 | H 4 | Y C A I I a | Y C A I I b | 4 5 6 | 6 0 7 | 5 7 6 | 5 7 0 | C D Y a | C D Y b | 4 4 2 | 4 3 8 | 4 2 5 | 4 4 4 | 4 4 6 | 5 3 1 | 5 7 8 | 3 9 5 S 1 a | 3 9 5 S 1 b | 5 9 0 | 5 3 7 | 6 4 1 | 4 7 2 | 4 0 6 S 1 | 5 1 1 | 4 1 3 a | 4 1 3 b | 5 5 7 | 5 9 4 | 4 3 6 | 4 9 0 | 5 3 4 | 4 5 0 | 4 8 1 | 5 2 0 | 6 1 7 | 5 6 8 | 4 8 7 | 5 7 2 | 6 4 0 | 4 9 2 | 5 6 5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FETYY | Hullinger | Boniswyl, Aargau, Switzerland | 12 | 23 | 14 | 10 | 14 | 18 | 11 | 15 | 13 | 13 | 11 | 29 | 14 | 8 | 9 | 11 | 11 | 26 | 15 | 21 | 31 | 12 | 12 | 15 | 16 | 11 | 10 | 19 | 21 | 17 | 15 | 17 | 18 | 36 | 37 | 12 | 9 | 12 | 11 | 13 | 11 | 7 | 14 | 15 | 8 | 11 | 10 | 8 | 10 | 9 | 17 | 17 | 15 | 10 | 12 | 12 | 15 | 9 | 23 | 21 | 12 | 11 | 14 | 11 | 12 | 12 | 12 |
BRANCH: L26
AGE: TO BE DETERMINED
LOCATION OF ORIGIN: WEST ASIA
Today, the distribution and frequency of this lineage’s members echoes the origins of their ancestor. It is 10 to 11 percent of the male population of Tunisia. It is about 5 percent of modern Macedonian male lineages. It is 4 to 5 percent of the male population of Cyprus. It is 1 to 2 percent of male lineages in Switzerland. Geneticists have found this lineage at trace frequencies of less than 1 percent through most of Western and Central Europe.
Note: This branch is not accompanied by a major movement on the map, and research on this branch is continuing.
The map below shows the path and cocentrations of our male ancestors and their descendants.
J-FGC9961/FGC9876/J-F761 Geographic Distribution
J-FGC9961/FGC9876/J-F761 Geographic Distribution
Points of origin for Geno2 SNPJ-F761, a subclade of J-L25/J2a4h. Equivalent SNP's FGC9961, FGC9962, FGC9876. Subclade resides downstream of J-F3133
Genetic "Cousins"
Our ancestors got around.
Click to go to the site
Phylogenetic Tree of Haplogroup J2
Two main subclades divide haplogroup J2: J2a (M410, L152, L212/PF4988, L559/PF4986) and J2b (M12, M102, M221, M314).
- J2a1-M47 is found at low frequency (1-5%) in Anatolia, Georgia, Iran, Iraq and Gulf states.
- J2a1-M67 is the most common subclade in the Caucasus (Vainakhs, Ingushs, Chechens, Georgians, Ossetians, Balkars) and in the Levant (Lebanese, Jews). It is also common in western India, the Arabian Peninsula, Anatolia (esp. north-west), Greece (esp. Crete), Italy (esp. Marche and Abruzzo) and Iberia. M67 was probably a major Neolithic lineage expanding from the Fertile Crescent to Greece to the west and the Indus valley to the east.
- J2a1-M68 a minor subclade found in Iraq and India.
- J2a1-M319 has been found chiefly in Greece (esp. in Crete) and Italy, and at low frequencies around Western Europe (perhaps diffused by the Romans).
- J2a1-M339 is a very minor Anatolian subclade.
- J2a1-M419 is a minor subclade detected in northern Iran.
- J2a1-P81 is a very minor Anatolian subclade.
- J2a1-L24 is the most widespread subclade of J2a, with a distribution ranging from the Middle East to Europe, North Africa and South Asia.
- J2a1-M158 has been found in Anatolia, Iberia, Pakistan and India.
- J2a1-L84 is a minor subclade detected in the Balkans.
- J2a1-L25 is the main branch of L24 and is subdivided in many subclades.
- J2a1-F3133 is found in Anatolia, Syria, Iran, Central Asia and Saudi Arabia.
- J2a1-F761 is the Western European subclade of F3133, found in Italy, France, the Benelux and England.
- J2a1-L192.2 is found in Anatolia, Iran and Kerala (India). It has also been found in Tunisia (M'saken).
- J2a1-PF4888 is found in the Middle East and among Ashkenazi Jews (F659 subclade: Katz and Cohen).
- J2a1-Z387 and its main subclade L70 (DYS445≤7) are found throughout continental Europe as well as in the Middle East at lower frequency.
- J2a1-PF5169 is a rare subclade that has been found in Saudi Arabia, Switzerland, southern Germany and England.
- J2a2-PF7381 is found at low frequency in southern and Eastern Europe and in the Caucasus.
- J2b1-M205 is found mostly in the southern Balkans and Anatolia.
- J2b2-M241 is found mostly in south-east, central and Eastern Europe and in India.
Some Great New Info Shared by Robert H.A. Sanders


Y-DNA Haplogroup J2 M172 - Roman Empire
While it has been said before in forums, chats and websites on the internet that the J2 Frequency Map for Europe shows a resemblance to the borders of the Roman Empire, i have never seen an actual image comparing the two. I decided to make one for myself and upload it here. I used the current (2012) Eupedia.com J2 Frequency map and an image of the Roman Empire at its largest extent (about 117 AD). Eupedia.com tells us; "Romans surely helped spread haplogroup J2 across its borders, judging from the distribution of J2 within Europe (frequency over 5%) which bears an uncanny resemblance to the borders of the Roman Empire."
Rober H. A. Sanders
Our J2 Haplogroup passed from father to expanded in a pattern similar to the expansions of the Phoencians.
Y-DNA Haplogroup J2 M172, quotes & links. List compiled by R.H.A. Sanders, 2013. ]-----------------------
"In human genetics, Haplogroup IJ is a human Y-chromosome DNA haplogroup. Haplogroup IJ is a descendant branch of Haplogroup F-L15 which in turn derives from the greater Haplogroup F. Descendants are Haplogroup I and Haplogroup J." Haplogroup IJ en.wikipedia.org/wiki/
"In human genetics, Haplogroup IJ is a human Y-chromosome DNA haplogroup. Haplogroup IJ is a descendant branch of Haplogroup F-L15 which in turn derives from the greater Haplogroup F. Descendants are Haplogroup I and Haplogroup J." Haplogroup IJ en.wikipedia.org/wiki/
"Various episodes of population movement have affected southeast Europe, and the role of the Balkans as a longstanding gateway to Europe from the Near East is illustrated by the phylogenetic unification of Hgs I and J by the basal M429 mutation. This evidence of common ancestry suggests that ancestral IJ-M429* Y chromosomes probably entered Europe through the Balkan route sometime before the Last Glacial Maximum."
Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. unipv.eu/on-line/
"J-M172 can be classified as Greco-Anatolian, Mesopotamian and/or Caucasian and is linked to the earliest indigenous populations of Anatolia. It was carried by Bronze Age immigrants to Europe, and ultimately descends from the Cro-Magnon population (IJ-M429 Y-DNA) that emerged in Southwest Asia around 35,000 years ago"
Wikipedia.org - Haplogroup J2 M172.
"A 2004 study by Semino et al. contradicted this study, and showed that Italians in North-central regions (like Tuscany and Emilia-Romagna) had a higher concentration of J2 than their Southern counterparts. North-central had 26.9% J2, whereas Calabria (a far Southern region) had 20.0%, Sardinia had 9.7% and Sicily had 16.7%. This could be because of the ancient Etruscans, who some think originated in the Near East." Wikipedia.org - Genetic History of Italy. en.wikipedia.org/wiki/
"From these comparisons, we found that haplogroup J2, in general, and six Y-STR haplotypes, in particular, exhibited a Phoenician signature that contributed > 6% to the modern Phoenician-influenced populations examined."
"From these comparisons, we found that haplogroup J2, in general, and six Y-STR haplotypes, in particular, exhibited a Phoenician signature that contributed > 6% to the modern Phoenician-influenced populations examined."
Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean.
http://www.cell.com/AJHG/
The Neolithic control section shows nonsignificant results across all haplogroups, except for a significant J2 result in one test. The Phoenician-colony test results highlight only one haplogroup, J2, which consistently scores significantly in all three tests across the range of colonization sites. However, this haplogroup also scores significantly in Greek tests (as do some additional haplogroups), suggesting that the same haplogroup could have been spread by several expansions, which is unsurprising considering its frequency in the Eastern Mediterranean but implies that higher phylogenetic resolution is required for identification of Phoenician-specific signals."
The Neolithic control section shows nonsignificant results across all haplogroups, except for a significant J2 result in one test. The Phoenician-colony test results highlight only one haplogroup, J2, which consistently scores significantly in all three tests across the range of colonization sites. However, this haplogroup also scores significantly in Greek tests (as do some additional haplogroups), suggesting that the same haplogroup could have been spread by several expansions, which is unsurprising considering its frequency in the Eastern Mediterranean but implies that higher phylogenetic resolution is required for identification of Phoenician-specific signals."
Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean.
http://www.cell.com/AJHG/
"In addition to Hg J-M410, Hg G-P15 chromosomes, which are also common in Anatolia,29 have been implicated in the colonization and subsequent expansion of early farmers in Crete, the Aegean and Italy.38,46 - 48 Earlier studies have concluded that the J-M410 sub-clades, J-DYS445-6 and J-M67, are linked to the spread of farming in the Mediterranean Basin,38,47 with a likely origin in Anatolia.29 Interestingly, J-DYS445-6 and J-M92 (a sub-lineage of M67), both have expansion times between 7000 and 8000 years ago (Table 1), consistent with the dating of the arrival of the first farmers to the Balkans." Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. http://www.unipv.eu/on-line/
"In addition to Hg J-M410, Hg G-P15 chromosomes, which are also common in Anatolia,29 have been implicated in the colonization and subsequent expansion of early farmers in Crete, the Aegean and Italy.38,46 - 48 Earlier studies have concluded that the J-M410 sub-clades, J-DYS445-6 and J-M67, are linked to the spread of farming in the Mediterranean Basin,38,47 with a likely origin in Anatolia.29 Interestingly, J-DYS445-6 and J-M92 (a sub-lineage of M67), both have expansion times between 7000 and 8000 years ago (Table 1), consistent with the dating of the arrival of the first farmers to the Balkans." Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. http://www.unipv.eu/on-line/
"Regarding Hg J-M12/M102, which is discernable from India to Europe, the M12/M102* chromosomes display a very high YSTR diversity, whereas on the other hand, the J-M241 sub-lineage has low diversity in the Balkans, indicating different demographic histories. Although Hg J-M241 shows high variance in India, its place of origin is still uncertain. As J-M241 has older expansion times in Sicily, Apulia and Turkey, it may have arrived in the Balkans from elsewhere." Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. http://www.unipv.eu/on-line/
"The PC analysis, from the perspective of population Hg frequencies, reveals a tight cluster of populations not comprising southern Balkan and Caucasian groups. Common to this cluster are lower frequencies of Hgs, G-M201 and J-M410, and higher frequencies of Hgs, I-M423, E-V13 and J-M241. Whereas the first two are primarily Middle Eastern Hgs and have been shown to be associated with the early Neolithic colonization of Crete, Italy and southern Caucasus, I-M423, E-V13 and J-M241, in spite of parallel Balkan patterns of distribution, have clearly different origins." Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. http://www.unipv.eu/on-line/
"The majority of the Balkan Hg J Y chromosomes belong to the J-M172 sub-Hg and range from 2% to 20%. Both its main branches, J-M410 and J-M12/M102*, were observed; although the first is scattered in different sub-clades (J-M67, J-M92 and J-DYS445-6) with distinct local patterns, the second is most represented by J-M241." Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. http://www.unipv.eu/on-line/
"Occurrence of J2-M172 Y-chromosomes in Tuscany has been related to the Etruscan heritage of the region." Uniparental Markers of Contemporary Italian Population Reveals Details on Its Pre-Roman Heritage. http://www.plosone.org/
"The majority of the Balkan Hg J Y chromosomes belong to the J-M172 sub-Hg and range from 2% to 20%. Both its main branches, J-M410 and J-M12/M102*, were observed; although the first is scattered in different sub-clades (J-M67, J-M92 and J-DYS445-6) with distinct local patterns, the second is most represented by J-M241." Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. http://www.unipv.eu/on-line/
"Occurrence of J2-M172 Y-chromosomes in Tuscany has been related to the Etruscan heritage of the region." Uniparental Markers of Contemporary Italian Population Reveals Details on Its Pre-Roman Heritage. http://www.plosone.org/
"There is a distinct association of ancient J2 civilisations with bull worship. The oldest evidence of a cult of the bull can be traced back to Neolithic central Anatolia, notably at the sites of Çatalhöyük and Alaca Höyük. Bull depictions are omnipresent in Minoan frescos and ceramics in Crete. Bull-masked terracotta figurines and bull-horned stone altars have been found in Cyprus (dating back as far as the Neolithic, the first presumed expansion of J2 from West Asia)." The Sacred Bull.
http://aratta.wordpress.com/
"The most frequent haplogroups among the current population on Crete were: R1b3-M269 (17%), G2-P15 (11%), J2a1-DYS413 (9.0%), and J2a1h-M319 (9.0%). They identified J2a parent haplogroup J2a-M410 (Crete: 25.9%) with the first ancient residents of Crete during the Neolithic (8500 BCE - 4300 BCE) suggesting Crete was founded by a Neolithic population expansion from ancient Turkey/Anatolia." The Minoans, DNA and all. http://
"The most frequent haplogroups among the current population on Crete were: R1b3-M269 (17%), G2-P15 (11%), J2a1-DYS413 (9.0%), and J2a1h-M319 (9.0%). They identified J2a parent haplogroup J2a-M410 (Crete: 25.9%) with the first ancient residents of Crete during the Neolithic (8500 BCE - 4300 BCE) suggesting Crete was founded by a Neolithic population expansion from ancient Turkey/Anatolia." The Minoans, DNA and all. http://
"We reconstructed the genetic structure of the Levantines and found that a pre-Islamic expansion Levant was more genetically similar to Europeans than to Middle Easterners." Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture.
"23andMe has a Y chromosome marker on its custom chip, rs34126399, which captures the spread of agriculture from the Near East to Europe. The G state at rs34126399 is found in most individuals carrying paternal haplogroup J2a, whose origin can ultimately be traced to Turkey 15,000 to 20,000 years ago." The Origin of Farming in Europe: A View from the Y Chromosome. http://blog.23andme.com/
"The authors found a weak - but significant - genetic signature among their samples that could not be explained by chance. Many of the samples belonged to a very specific branch of haplogroup J2, which the authors believe points back to distinct migrations by Phoenician traders from the Middle East into Europe and North Africa more than 3,000 years ago." Ripples in the Mediterranean: Tracing the Genetic Origins of the Phoenicians.
"R1b3 frequency was found to be higher in the northern part of the country, while the Y-chromosome haplogroups G and E3b1, J2 and I(xI1b2) frequencies were higher in the south and in the central part of the country, respectively."
Uniparental Markers of Contemporary Italian Population Reveals Details on Its Pre-Roman Heritage. http://www.plosone.org/
"Bulgarian DNA profile is congruent with those described for most European populations. Almost the entire Bulgarian mtDNA pool is made up of West Eurasian lineages, with just 0.9% of Eastern Asian lineages. It is a similar picture from Y-chromosome haplogroups. About 80% of the total genetic variation in Bulgarians falls within haplogroups E-M35, I-M170, J-M172, R-M17 and R-M269, all found elsewhere in Europe." Bulgarians. http://en.wikipedia.org/wiki/
"Haplogroup J2 is most common in Southern Europe, Anatolia and the Caucasus, were it may have originated 18.000 years ago. It appears to have spread into Europe in a number of waves over the course of millennia." 23andme.com, 2013. https://www.23andme.com/
"It has been plausibly suggested that M172 may be associated with the arrival of neolithic farmers from the Fertile Crescent who were the probable predecessors of the Indo-European society which later emerged in western Asia, a "hypothetical" society whose culture and language greatly influenced prehistoric peoples from India to Ireland." Genetics & Anthropology in Sicily.
"Haplogroups E1b1b and J in Europe are regarded as markers of movements from southeastern Europe to northwestern and therefore as a potential markers of introduced technology such as farming." Genetic history of the British Isles. http://en.wikipedia.org/wiki/
"J-M172, which occurs as frequently as J-M267 in some Middle Eastern populations, is the more prevalent in Europe." Origin Diffusion and Differentation Y-Chromosome Haplogroups E and J. http://www.ncbi.nlm.nih.gov/
"The J-M67*, JM92, and J-M102 representatives reflect more distinctive origins and dispersal patterns. Whereas J-M67* and J-M92 show higher frequencies and variances in Europe (0.40 and 0.32, respectively) and in Turkey (0.32 and 0.30, respectively [Cinniog˘ lu et al. 2004]) than in the Middle East (0.17 and 0.09, respectively), J-M12(M102) shows its maximum frequency in the Balkans." Origin Diffusion and Differentation Y-Chromosome Haplogroups E and J. http://www.ncbi.nlm.nih.gov/
"The diversity within J2 is lower in the Middle East (0.43 ±0.11) compared with both Turkey (0.60±0.07) and the European locations (0.67±0.02)." Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. http://www.familytreedna.com/
"Based on previously published data (Scozzari et al. 2001; Di Giacomo et al. 2004; Semino et al. 2004; Marjanovic et al. 2005), we observed that another haplogroup, J-M12, shows a frequency distribution within Europe similar to that observed for E-V13."
Tracing past human male movements in northern/eastern Africa and western Eurasia. http://mbe.oxfordjournals.org/
We favor the emergence of J2f1 in the Aegean area, possibly during the population expansion phase also
detected by Malaspina et al. (2001) and coincident with the expansion of the Greek world to the European coast of the Black sea."
Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. http://www.familytreedna.com/
"The M172 marker defines a major subset of M304, which arose from the M89 lineage. It is found today in North Africa, the Middle East, and southern Europe. In southern Italy it occurs atfrequencies of 20 percent, and in southern Spain, 10 percent of the population carries this marker. Both M304 and its subgroup M172 are found at a combined frequency of around 30 percent amongst Jewish individuals. The early farming successes of these lineages spawned population booms and encouraged migration throughout much of the Mediterranean world." National Geographic - Genographic Project, 2011. https://genographic.
"J2-M172 is more prevalent in Europe where at least five different lineages can be traced--J2e*-M102, J2e1-M241, J2*-M172, J2f*-M67, and J2f1-M92 (fig. 2, Semino et al. 2004)." High-Resolution Phylogenetic Analysis of Southeastern Europe Traces Major Episodes of Paternal Gene Flow Among Slavic Populations. http://mbe.oxfordjournals.org/
"The finding that five major haplogroups (E3b1, I1-P37 (xM26), J2, R1a, and R1b) comprise more than 70% of SEE total genetic variation is consistent with the typical European Y chromosome gene pool." Implications of the role of Southeastern Europe in the origins and diffusion of major Eurasian paternal lineages. http://arheologija.ff.uni-lj.
"Anthropologist Carleton S. Coon is quoted as saying The Iraqi population is without doubt much the same today as it was in Sumerian, Akkadian, Assyrian and Babylonian times. The Iraqi people are a Caucasian people. It has been found that Y-DNA Haplogroup J2 originated in northern Iraq (Ancient Assyria)." Wikipedia.com - Archeogenetics of the Near East. http://en.wikipedia.org/wiki/
"Romans surely helped spread haplogroup J2 across its borders, judging from the distribution of J2 within Europe (frequency over 5%) wich bears an uncanny resemblance to the borders of the Roman Empire." Eupedia.com , 2013. http://www.eupedia.com/europe/
"It has been proposed that haplogroup subclade J-M410 was linked to populations on ancient Crete by examining the relationship between Anatolian, Cretan, and Greek populations from around early Neolithic sites in Crete." Wikipedia.org - Haplogroup J2 M172.
http://en.wikipedia.org/wiki/
"The world`s maximum concentrations of J2a is in Crete (32% of the population). The subclade J2a4d (M319) appears to be native to Crete." Eupedia.com 2013. http://www.eupedia.com/europe/
"The world`s maximum concentrations of J2a is in Crete (32% of the population). The subclade J2a4d (M319) appears to be native to Crete." Eupedia.com 2013. http://www.eupedia.com/europe/
"Haplogroup J-M12 was associated with Neolithic Greece (ca. 8500 - 4300 BCE) and was reported to be found in modern Crete (3.1%) and mainland Greece (Macedonia 7.0%, Thessaly 8.8%, Argolis 1.8%) (King 2008)." Wikipedia.org - Haplogroup J2 M172.
"When looking at the diffusion of Haplogroup J2a, M410, westward into Europe, one aspect of this westward spread becomes quite clear. M410+ ancestors used a maritime and coastal route to move west."
M172 Blog - Pronounced Westward Maritime Diffusion of J2a (M410), 2008.
"Dr. King also notes an interesting correlation with a subclade of Haplogroup J2, M67, and place names in the Aegean, Balkans and Italy while citing a deeper origin for subclade M67 in Northern Syria or southern Anatolia. The age and spread of M67 seems associated with proto-greek substratum in the Aegean."
M172 Blog - Neolithic Migrations in the Near East and Aegean, 2009.
"Quite a few ancient Mediterranean and Middle Eastern civilisations flourished in territories where J2 lineages were preponderant. This is the case of the Hattians, the Hurrians, the Etruscans, the Minoans, the Greeks, the Phoenicians (and their Carthagian offshoot), the Israelites, and to a lower extend extent also the Romans, the Assyrians and the Persians. All great seafaring civilisations from the middle Bronze Age to the Iron Age were dominated by J2 men." Eupedia.com - Haplogroup J2.
"While noting that multiple haplogroups are likely involved in the spread of languages through the middle east, Dr. King noted a correlation between very old Middle Eastern languages of uncertain origin and Haplogroup J2 while at the same time theorizing that Haplogroup J1 may have been involved in spreading Semitic languages through the region. These old languages possibly linked to J2 are known to have existed in Mesopotamia and the Northern Levant and this substratum is sometimes referred to as "Banana" languages due to their syllabic duplication." M172 Blog - Neolithic Migrations in the Near East and Aegean, 2009.
"Both E-V13 and J-M12 have also been used in studies seeking to find evidence of a remaining Greek presence in Afghanistan and Pakistan, going back to the time of Alexander the Great." Wikipedia.org - Haplogroup E V-68. http://en.wikipedia.org/wiki/
"A genetic study published led by Firasat (2007) on Kalash individuals found high and diverse frequencies of :Haplogroup L3a (22.7%), H1* (20.5%), R1a (18.2%), G (18.2%), J2 (9.1%), R* (6.8%), R1* (2.3%), and L* (2.3%).[39] Haplogroup L, Haplogroup H, and Haplogroup R1a are thought to have originated from prehistoric South Asia." Wikipedia.org - Kalash People.
http://en.wikipedia.org/wiki/
"The sister clade to J2a-M410 is J2b-M12. In India and Pakistan, all J2b members comprise the J2b2-M241 derivative HG." Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists.
"The sister clade to J2a-M410 is J2b-M12. In India and Pakistan, all J2b members comprise the J2b2-M241 derivative HG." Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists.
http://www.ncbi.nlm.nih.gov/
"Lastly, HG J2b2-M241-related microsatellite variance is higher in Uttar Pradesh near the border of Nepal. It should be noted that numerous Mesolithic sites have been observed in this region (Kennedy 2000)." Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists.
"Lastly, HG J2b2-M241-related microsatellite variance is higher in Uttar Pradesh near the border of Nepal. It should be noted that numerous Mesolithic sites have been observed in this region (Kennedy 2000)." Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists.
"J2b has a quite different distribution from J2a. J2b seems to have a stronger association with the Chalcolithic cultures of Southeast Europe, and is particulary common in the Balkans, Central Europe and Italy, which is roughly the extent of the European Copper Age culture" Eupedia.com 2013 http://www.eupedia.com/europe/
"J2a is also present in Egypt which was conquered by Macedonian Greeks, as well as Iran, but drops to a small frequency in India, and is there limited to the upper castes. This may reflect its presence in the ancient Indo-Aryans and its survival in the Brahmin caste, or alternatively may be the result of intermarriage between the Bactrian Greek aristocracy and high-class Hindus" Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"Haplogroup J2a-M410 is confined to upper caste Dravidian and Indo-European speakers, with little occurrence in the middle and lower castes." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"The J2 clade is nearly absent among Indian tribals, except among Austro-Asiatic speaking tribals (11%). Among the Austro-Asiatic tribals, the predominant J2b2 hg occurs only in the Lodha." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"Haplogroup J2a-M410 is confined to upper caste Dravidian and Indo-European speakers, with little occurrence in the middle and lower castes." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"The J2 clade is nearly absent among Indian tribals, except among Austro-Asiatic speaking tribals (11%). Among the Austro-Asiatic tribals, the predominant J2b2 hg occurs only in the Lodha." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"One fourth of the Vlach people (isolated communities of Romance language speaking peoples in the Balkans) belong to J2, which, combined to the fact that they speak a language descended from latin, suggests that they could have had a greater part of Roman (italian) ancestry than other ethnic groups in the Balkans." Eupedia.com 2013. http://www.eupedia.com/europe/
"The remaining two haplogroups, J2 and E3b, exhibit spotty frequencies in Russians, expected for low-frequency haplogroups. The haplogroups might have arrived to Russia alongside I1b from the Balkans, in which the two are frequent." Two Sources of the Russian Patrilineal Heritage in Their Eurasian Context. http://www.sciencedirect.com/
"Sicily has one of the highest frequencies of Haplogroup J2 (M172) in the mediterranean. J2-M172 made up 33% of the Y chromosome signatures on the island and was non-randomly distributed occurring at higher frequencies in the eastern areas of the island. This distinction was evident in the subclades, M67 and M92, which have previously been linked to Greek and proto-greek colonization. Both M67 and M92 were twice as frequent on the eastern portion of Sicily which displays more archaelogical traces from the Greek classic era. Even the paragroup of undistinguished J2 haplotypes (M172) was more than twice as frequent in Eastern Sicily." M172 Blog - Y Chromosomes of Sicily, 2008. http://m172.blogspot.nl/2008/
"In turn, two distinct haplogroups, J2a1h-M319 and J2a1b1-M92, had demographic properties consistent with Bronze Age expansions to Crete, arguably from NW/W Anatolia and Syro-Palestine, while a later mainland (Mycenaean) contribution to Crete was indicated by the presence of of V13." Implications of the role of Southeastern Europe in the origins and diffusion of major Eurasian paternal lineages. http://arheologija.ff.uni-lj.
"The Neolithic component in the SEE paternal gene pool is most clearly marked by the presence of the J-M241 (more frequent in the Southern Balkans) lineage, and its expansion signals associated with Balkan microsatellite variation correlate with the Neolithic period." Implications of the role of Southeastern Europe in the origins and diffusion of major Eurasian paternal lineages. http://arheologija.ff.uni-lj.
"Within India, J2a is more common among the upper castes and decreases in frequency with the cast level." Eupedia.com, 2013 http://www.eupedia.com/europe/
"There is a distinct association of ancient J2 civilisations with bull worship" Eupedia.com 2013 http://www.eupedia.com/europe/
"In all likelihood, J2a1 originated before the ethnogenesis of the Greeks, and may be associated with multiple population movements from the Greek-Balkan region. However, I believe that it makes better sense to view it as a Balkan-Greek clade than a West-Asian one." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"In all likelihood, J2a1 originated before the ethnogenesis of the Greeks, and may be associated with multiple population movements from the Greek-Balkan region. However, I believe that it makes better sense to view it as a Balkan-Greek clade than a West-Asian one." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"The higher frequency of J2 in southern Italy and Sicily compared to northern Italy, is also explained by this theory, as these regions were colonized by Greeks, whereas northern Italy was not." Dienekes Anthropology Blog, 2005. http://dienekes.blogspot.nl/
"The propagation of J2b and E V-13 correspond roughly to the ancient Greek and Roman spheres of influence." Eupedia.com, 2013. http://www.eupedia.com/europe/
"The ancient Greeks and Phoenicians were the main driving forces behind the spread of J2 around the western and southern Mediterranian" Eupedia.com, 2013. http://www.eupedia.com/europe/
"The propagation of J2b and E V-13 correspond roughly to the ancient Greek and Roman spheres of influence." Eupedia.com, 2013. http://www.eupedia.com/europe/
"The ancient Greeks and Phoenicians were the main driving forces behind the spread of J2 around the western and southern Mediterranian" Eupedia.com, 2013. http://www.eupedia.com/europe/
"Wine making spread to Crete during the Minoan period and then later to Italy with the Etruscans and to Iberia with the Phoenicians. It was an integral component of the economy and social culture of the proto-greek civilizations and the phoenicians who both went on to settle other mediterranean coastal regions. And tracing the spread of Viticulture from its origins to its spread before the Roman period, we can see te highest levels of Haplogroup J2 today correlate with the geographical centres of all these civilizations. While viticulture may not represent the first wave of M172 migrants to Europe, M172 certainly played a strong role in bringing Viticulture to Europe with such civilizations as the Minoans, Greeks and Phoenicians."
M172 Blog - Correlations in the spread of Viticulture and Haplogroup J2, 2008.
"Haplogroup J2b-M12 was frequent in Thessaly and Greek Macedonia while haplogroup J2a-M410 was scarce. Alternatively, Crete, like Anatolia showed a high frequency of J2a-M410 and a low frequency of J2b-M12" Differential Y-chromosome Anatolian Influences on the Greek and Cretan Neolithic, 2008. http://www.atlascom.gr/
"Di Giacomo stressed the role of post-Neolithic migratory phenomenon, specifically that of the Ancient Greeks, as also being important in the dispersal of haplogroup J-M172." Wikipedia.org - Haplogroup J2 M172. http://en.wikipedia.org/wiki/
"Di Giacomo stressed the role of post-Neolithic migratory phenomenon, specifically that of the Ancient Greeks, as also being important in the dispersal of haplogroup J-M172." Wikipedia.org - Haplogroup J2 M172. http://en.wikipedia.org/wiki/
"According to Di Giacomo's (2004) study, the high diversity of haplogroup J2 in Turkish and southern European populations suggests that this branch of haplogroup J originated around the Aegean, not the Middle East. Additionally, it appears that much of J2 was confined to the coastal Mediterranean areas, indicating that maritime trade, rather than earlier Neolithic agricultural expansions, may have helped spread J2 throughout the Mediterranean world." A reassessment of Jewish DNA Evidence.
"The two haplogroups most strongly associated with Albanian people (E-V13 and J2b) are often considered to have arrived in Europe from the Near East with the Neolithic revolution or late Mesolithic, early in the Holocene epoch. From here in the Balkans, it is thought, they spread to the rest of Europe." Wikipedia.org - Origins of the Albanians. http://en.wikipedia.org/wiki/
"Y-Dna haplogroups are found at the following frequencies in Malta : R1 (35.55% including 32.2% R1b), J (28.90% including 21.10% J2), I (12.20%), E (11.10% including 8.9% E1b1b), F (6.70%), K (4.40%), P (1.10%).[23] Haplogroup R1 , E1b1b, J2 and I are typical in European populations. J1, K, F haplogroups consist of lineages with differential distribution within Middle East, North Africa and Europe. The low percentages of J1 are similar to the Sicilian population, suggesting common ancestry with Sicilians and negligible genetic input from both North Africa and the Middle East." Wikipedia.org - Maltese People. http://en.wikipedia.org/wiki/
"Timur Serdar and Demircin Sema authored a recent study on the Y chromosomes of Antalya, which is located on the southern coast of Anatolia. Haplogroup J2 was most frequent in this study of 75 unrelated males found at a frequency of 26.6%. The J2 data was consistent with an earlier study by Cinnioglu et al which found 24% J2 in southern Anatolia. Haplogroup T (K* in the study) was next most frequent at 13.3% and this data differed from Cinnioglu's data which found only 3.3% Haplogroup K in southern Anatolia. The first record of Antalya was as Attalia, a greek city founded approximately 150BC by Attalos II, King of Pergamon."
M172 Blog - Haplogroup J2, M172 in Antalya, Turkish Republic, 2009.
"In 2004, two geneticists educated at Harvard University and leading scientists of the National Geographic Genographic Project, Dr. Pierre Zalloua and Dr. Spencer Wells, identified "the haplogroup of the Phoenicians" as haplogroup J2, with avenues open for future research." Familypedia.com - Phoenicia. http://familypedia.wikia.com/
"Di Giacomo's (2004) study emphasized that J2 is "Mediterranean" or "Aegean" rather than "Semitic" in character. It is found predominately in northern Mediterranean and Turkish populations, differentiating the Aegean area from the Middle East in its haplogroup J results. Going further, the researchers maintained that certain sub-clades of J2 appear to have originated well after the beginning of the Neolithic revolution and around the Aegean, spreading out to the rest of Europe during the expansion of the Greek world. It is this final idea - that much of J2 is European in origin rather than Middle Eastern - that complicates the interpretation of Jewish J2 results. Sub-clade J-M102* originated in the southern part of the Balkans and is generally absent in Middle Eastern populations (Semino et al. 2004). Ashkenazim have a 1.2% frequency of J-M102 and Sephardim have 2.4%. These results argue in favor of European gene flow into the Jewish community." A reassessment of Jewish DNA Evidence.
"The strong western (-0.82) but weak southern (-0.37) orientation of J2 is unexpected if J2 came to Germany from the Balkans, but is consistent with a maritime mode of propagation of this haplogroup. Interestingly, the J2 frequencies in French (5-17.3%), Dutch (6.2%) and Belgian (5.0%) samples all exceed the German average (4.0%), so they are probably consistent with this interpretation. We really need to differentiate between J2a and J2b clades in this area, since J2a may hold the promise of reflecting maritime colonization (as its high frequency in coastal and island southern Europe suggests) or Roman descendants, while J2b may hold the signal of an expansion out of the Balkan area." Dienekes Anthropology Blog, 2008. http://dienekes.blogspot.nl/
"In deze studie wordt een tot nu toe niet bekende concentratie van de haplogroepen J in oostelijk Brabant aangetroffen. Het betreft hier voornamelijk J2 en oude, gevestigde families. Het is niet onmogelijk dat hier sprake is van families van nazaten van ´Romeinse´soldaten. Ook in het grensgebied van Engeland en Schot+and is een concentratie van J2 families gevonden. In dat gebied zijn ook resten van tempels van de Iraanse god Mitras bekend en blijkt er een boogschuttereenheid uit het Midden/Oosten gelegerd geweest te zijn. Romeinse aanwezigheid in het land van Cuijck en aan de randen van de Peel (de naam is afkomstig van de naam die de Romeinen aan het gied gaven: Locus Paludosus ofwel moerassige streek) is bekend, zoals ook landmetingen van militairen die zich er gevestigd hadden en landbouw bedreven." Project Genetische Genealogie in Nederland. http://www.barjesteh.nl/
"IE-speaking Iranians have largely the same haplogroups as Arabs, but a much higher representation of haplogroup J2 compared to J1. The converse is true for all Arabs except the Lebanese. But, we do know, that even in Lebanon itself, Muslims have a higher J1/J2 ratio than Christians, and Islam was the main vehicle of Arabization in the region. The Christians are descended from the pre-Arab Byzantine Greco-Aramaic populations." Coastal-inland differences in Y chromosomes of the Levant.
"Results derived from analysis of the non-recombining portion of the Y- chromosomes (NRY) produced, at least initially, similar gradients to the classic demic diffusion hypothesis. Two significant studies were Semino 2000 and Rosser 2000, which identified Haplogroup J2 and E1b1b (formerly E3b) as the putative genetic signatures of migrating Neolithic farmers from Anatolia, and therefore represent the Y-chromosomal components of a Neolithic demic diffusion. This association was strengthened when King and Underhill (2002) found that there was a significant correlation between the distribution of Hg J2 and Neolithic painted pottery in European and Mediterranean sites."
Wikipedia.com - Neolithic Europe. http://en.wikipedia.org/wiki/
"The Phoenicians, Greeks and Romans all contributed to the presence of J2a in Iberia. The particulary strong frequency of J2a and other Near Eastern haplogroups (J1, E1b1b, T) in the south of the Iberian peninsula, suggest that the Phoenicians and the Carthagians played a more decisive role than other peoples." Eupedia.com, 2013. http://www.eupedia.com/europe/
"The Phoenicians, Greeks and Romans all contributed to the presence of J2a in Iberia. The particulary strong frequency of J2a and other Near Eastern haplogroups (J1, E1b1b, T) in the south of the Iberian peninsula, suggest that the Phoenicians and the Carthagians played a more decisive role than other peoples." Eupedia.com, 2013. http://www.eupedia.com/europe/
"This lineage originated in the northern portion of the Fertile Crescent where it later spread throughout central Asia, the Mediterranean, and south into India. As with other populations with Mediterranean ancestry this lineage is found within Jewish populations."
FamilytreeDNA - SNP Certificate (Haplogroup J2). http://www.familytreedna.com/
"Because of your J2a1b association, it's possible your ancestors may have been members of populations which specifically expanded from Anatolia to the Greek island of Crete between 8500 and 4300 BC." Ancestry.com - Paternal Ancestry Certificate.
"The J2 haplogroup can be found in today's populations with notable frequency in Italy, Iberia, Turkey, Albania, Greece and even India, and most likely interacted with numerous cultures, including the Greeks and Romans."
Ancestry.com - Paternal Ancestry Certificate.
"Previously, the presence of Haplogroups J, E3b, and G among Jews was interpreted as additional evidence of Middle Eastern or Israelite ancestry in much the same fashion as the Cohanim Modal Haplotype. However, recent studies demonstrate that their origin is uncertain. Unfortunately, misinformation about these haplogroups continues to pervade the public and media. Haplogroup E3b is often incorrectly described as African, leaving a misimpression regarding the origin and complex history of this haplogroup. Haplogroup J2, as previously discussed, is often incorrectly equated with J1 and described as Jewish or Semitic, despite the fact that it is present in a variety of non-Jewish Mediterranean and Northern European populations."
A reassessment of Jewish DNA Evidence.
"The unique colonization pattern of the Phoenicians and the isolation of some of their colonies (Ibiza, Sardinia, Malta) have made it easy to identify their genetic signature. The Phoenician population was already very mixed 3000 years ago : E-V22, J1, J2, J2a4b, J2a4b1, G2a, R1a and R1b1a. E-V22 and R1b1a are quite specific to Levantines (Syrians, Lebanese, Druzes, Jews, Palestinians)."
Eupedia.com - Y-DNA haplogroups of ancient civilizations. http://www.eupedia.com/forum/
"From about 700 BCE, the Etruscans settled around Tuscany and the Greeks in southern Italy. Etruscans probably came from Palestine and brought haplogroups J1, J2 and E with them. The Greeks in Italy were Doric and brought J2, E, G2a and probably more R1b (see above). The Romans progressively absorbed the Etruscans and Italian Greeks and mixed with them. By the time of Julius Caesar Roman citizens were probably composed of 45% of R1b, 20% of J, 15% of E, 15% of G2a and 5% of I2a." Eupedia.com - Y-DNA haplogroups of ancient civilizations. http://www.eupedia.com/forum/
"Haplogroup J2 frequency has been correlated with aspects of the symbolic material culture of the Neolithic in Europe and the Near East (painted pottery and ceramic figurines) and sub-Haplogroups of J2 have also been associated with the Neolithic colonization of mainland Greece, Crete and southern Italy."
The coming of the Greeks to Provence and Corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean.
"Previous Y-chromosome genetic studies of Phoenician colonization have demonstrated that haplogroup J2 frequency was amplified in regions containing the Phoenician colonies of Iberia and North Africa in comparison to areas not containing Phoenician colonies."
The coming of the Greeks to Provence and Corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean.
"Many people new to Genetic Genealogy think the J2 haplogroup is synonymous with having male Jewish ancestry. One should note that having a J2 haplogroup assignment does not necessarily indicate Jewish ancestry. The J2 haplogroup is far more ancient than the Jewish religion and is found in many lines with Mediterranean region ancient ancestry. Another relatively more recent mode for J2's entry into some parts of Europe from the Mediterranean areas could have been the Roman Legions and Roman settlements."
Worldfamilies.net - Y-DNA Haplogroups. http://www.worldfamilies.net/
"Sicily is an island which had well-documented and not insignificant settlements by both Greeks and Phoenicians. Moreover, these settlements were geographically divided: Greeks in the East, Phoenicians in the West. It is in the East that J2 has its highest frequency, and not in the Phoenician West."
Dienekes Anthropology Blog, 2008. http://dienekes.blogspot.nl/
"From these comparisons, we found that haplogroup J2, in general, and six Y-STR haplotypes, in particular, exhibited a Phoenician signature that contributed > 6% to the modern Phoenician-influenced populations examined. Our methodology can be applied to any historically documented expansion in which contact and noncontact sites can be identified."
Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean.
"This blog is dedicated to those who carry the J2 "Y" DNA Haplogroup, with a focus on J2a4h2, also known as J-L25. Our "Y" Chromosome is inherited from father to son. Our paternal ancestors will also have the same signature. "J2 originated in northern Mesopotamia, and spread westward to Anatolia and southern Europe, and eastward to Persia and India. J2 is related to the Ancient Etruscans, (Minoan) Greeks, southern Anatolians, Phoenicians, Assyrians and Babylonians."
J2a4h Blog - J2a4h2 YHaplogroup J-L25 DNA.
http://j2a4h2.blogspot.nl/
J2a4h Blog - J2a4h2 YHaplogroup J-L25 DNA.
http://j2a4h2.blogspot.nl/
"Haplogroup J2 among Jews has been erroneously interpreted in the past as exclusively Israelite or Middle Eastern in origin. Among Ashkenazim, J2 occurs among 23.2% of the population, while Sephardim have 28.6% (Semino et al. 2004). While these percentages are nearly identical to Iraqi (22.4%) and Lebanese (25%) groups, they are also comparable to Greek (20.6%), Georgian (26.7%), Albanian (19.6%), Italian (20-29%), and to a lesser extent, French Basque (13.6%) populations (Semino et al. 2004)."
A reassessment of Jewish DNA Evidence.
"Haplogroup J is mostly found in South-East Europe, especially in central and southern Italy, Greece and Romania. It is also common in France, and in the Middle East. It is related to the Ancient Romans, Greeks and Phoenicians (J2), as well as the Arabs and Jews (J1). Subclades J2a and J2a1b1 are found mostly in Greece, Anatolia and southern Italy, and are associated with the Ancient Greeks." Romanian History and Culture. http://
"This lineage originated in the northern portion of the Fertile Crescent where it later spread throughout central Asia, the Mediterranean, and south into India. J2 is found in Britain, but rarely. It is most common in Eastern European countries, leading to speculation that it is either from gypsy background or, possibly, from Eastern European soldiers stationed in Britain during the Roman occupation in the first three centuries AD."
Scotland DNA Project.
"J2 has been traced back to the area between the Black Sea and the Caspian Sea that comprises territory in northwestern Iraq and Iran, eastern Turkey, Armenia, Azerbaijan and Georgia. One theory offered for the presence of J2 in northern England is the presence of Roman auxiliary soldiers stationed on Hadrian's Wall. It has been suggested that the original J2 ancestor of our Robson member may have been a Sarmatian horseman in the Roman legions." Robson/Robeson/Robison Family DNA Project (Haplogroup J2).
"The very name "Van Santen" means "from" or "of" Santen. The only Santen found (now known as "Xanten") is an ancient walled German town with a strong Roman history, at one time strategically located on the Rhine river (the river has since altered course somewhat). My genealogy is well-documented to the early 1500's from where my earliest known ancestor had settled by the 16th Century, near the mouth of the same Rhine river, and the furthest west one could have gone short of crossing the channel to England."
Van Santen DNA Project (Haplogroup J2).
"J2 - This haplogroup originated during the Neolithic in Central Asia, and spread across the Mediterranean and the Middle East. It may have been brought to Britain by prehistoric farmers, Greek or Phoenician traders and Sephardic Jews among the Normans and the Flemish - as well as by Roman troops and settlers."
Elliot (And border receivers) DNA Project (Haplogroup J2).
"Research conducted by the administrators of the Border Reivers DNA Project has identified numerous haplotypes in persons of British descent that show Haplotype 35 markers. Moreover, most of these haplotypes appear to originate from areas of Britain near the Antonine Wall, Hadrian's Wall and other places of Roman fortification or settlement. These areas include Galloway, Dumfries, Ayrshire and The Borders in Scotland, and Cumbria, Yorkshire, Lancashire, Shropshire and Staffordshire in England. Many of the Roman troops stationed in these areas came from Southeastern Europe or Western Asia. They included Sarmatians, Dacians, Goths, Syrians, Mesopotamians, Thracians and Anatolians. The Capelli study has shown that these areas also exhibit higher than average frequencies of haplogroups E3b and J2, neither of which is native to Britain. E3b is found most commonly in North Africa, Iberia, the Mediterranean and the Near East, and J2 occurs most frequently in the Near East, the Mediterranean and Western Asia. The fact that all three groups - E3b, J2 and Haplotype 35 - have a similar origin in territories of the Roman Empire, and occur at comparable frequencies in parts of Britain with a known history of Roman settlement, suggests that they arrived in Britain through the same means." Elliot (And border receivers) DNA Project (Haplogroup J2) - Haplogroup R1b (Haplotype 35). http://freepages.genealogy.
"In human genetics, Haplotype 35, also called ht35 or the Armenian Modal Haplotype, is a Y chromosome haplotype of Y-STR microsatellite variations, associated with the Haplogroup R1b. It is characterized by DYS393=12 (as opposed to the Atlantic Modal Haplotype, another R1b haplotype, which is characterized by DYS393=13). The members of this haplotype are found in high numbers in Anatolia and Armenia, with smaller numbers throughout Central Asia, the Middle East, the Balkans, the Caucus Mountains, and in Jewish populations. They are also present in Britain in areas that were found to have a high concentration of Haplogroup J, suggesting they arrived together, perhaps through Roman soldiers." Haplotype 35. http://en.wikipedia.org/wiki/
"Haplogroup J2 is the most common one today in the Middle East, but is found to some extent in adjacent regions. The bulk of J2 may have been brought to Britain by mercenaries recruited by the Romans."
Family Banks DNA Project (Haplogroup J2). https://sites.google.com/site/
"We now have two members of Haplogroup J1 and 6 members of Haplogroup J2 in the Fox Poject. This is a Mediterranean Haplogroup but exists all over Europe to some extent. The Romans are thought to have brought this Haplogroup to Britain." Fox FamilytreeDNA Project.
"The Greek and Phoenician presence also brought J2 into France and of course the Roman period also would have been a major contributor of Haplogroup J2 into what is today, France. So although, rare, Haplogroup J2 can be found in local populations throughout France and Spain."
Family Dugas, Haplogroup J2.
"The Mediterranean and Middle Eastern group consists of the the two Es, the G and the two J2s. Whit Athey and others have theorized that this group is associated with the Neolithic spread of agriculture from the Middle East into Europe. On the other hand, a more recent paper by Steven C. Bird argues for a Roman origin for J2 and E3b at least." Francis Surname Project. http://www.familytreedna.com/
"Sometime prior to that a Bretz ancestor must have come into Germany from the south, from Italy or Greece. Some researchers have suggested a family connection to Fabius Bretius, a Roman General, who came from the districts of Capua and Taranto in southern Italy to the town Trier, Germany around 224. Whether this connection is genuine or not, and it is impossible to know for sure, perhaps Roman conquests did bring Bretz DNA north 1,800 years ago as they did with so many other things. For some additional thoughts on the possible Latin origins of the family, also read the Genealogia Bretius."
Family Bretz, Haplogroup J2.
"Hoewel we tussen de periode van het ontstaan van onze haplogroep/subclade en het begin van de stamreeks de Sanders familiegeschiedenis niet met absolute zekerheid kunnen vaststellen ben ik van mening dat mede door het grote aantal indirecte aanwijzingen een redelijk nauwkeurig beeld van onze voorouders in die periode vastgesteld kan worden. Zo is het hoogstwaarschijnlijk dat onze voorouders in de tijd van het Romeinse rijk naar onze huidige geografische regio zijn gemigreerd. Zij waren waarschijnlijk Romeinse landbouwers en/of legionairs/veteranen van Italiaanse, Illyrische, Thracische of Griekse (Macedonische) afkomst. De indirecte relatie die we hebben met de van Santens geeft sterke aanwijzingen naar de plaats of regio Xanten, een oorspronkelijk Romeinse nederzetting die het middelpunt lijkt van de nederrijnse trojanenmythe. Deze Romeinse theorie sluit vervolgens perfect aan bij de eerstvolgende aanwijzing betreffende de afkomst van onze familie namelijk het Rooms Katholieke geloof."
Family Sanders, Haplogroup J2b2.
"My conclusion is that we are likely descendants of a Roman soldier serving in either Legio II or Legio XX. This Roman soldier was probably of Balkan (Macedonian, Illyrian, or Thracian) origin. It has been stated that ―The Romans surely helped spread haplogroup J2 within their borders, judging from the distribution of J2 within Europe (frequency more than 5 percent), which bear an uncanny resemblance to the borders of the Roman Empire. More research is needed to obtain a definitive answer but, in my opinion, we are most likely descendants of Roman soldiers recruited from the Balkans (Thrace, Macedonia, or Greece). The Hollywood movie ―King Arthur‖ (2004 by Touchstone Pictures) is a striking example of how our haplogroup could have arrived in Northwestern Europe in Roman times."
mr. R.H.A. Sanders, Haplogroup J2b2, Worden Family Newsletter August 2011. http://wordenfamilyassoc.org/
"J2 has been well studied and can be split into several clades but whose mode of individual distribution is not well understood. Many influences such as Greek and Roman would have played a part."
Wells Family DNA Project. http://www.rootsweb.ancestry.
"How our more recent ancestors ended up in England is still a mystery. It could have been a random migration of a single man. During the Roman occupation of Europe many people immigrated there from many areas of the world. Soldiers were sent there to serve military obligations. One of these might have been a man from the Mediterranean area. His descendants eventually took the surname Field and he might have been our earliest J2b2 ancestor." Genetic Journey of our branch of the Field family. http://www.luciefield.net/
"Haplogroup J2b is most common in the Middle East and reaches its highest percentages in Turkey. In Europe, the largest J2b populations are in Greece, Albania and Italy. This haplogroup is rare in Britain where it could represent remnants of eastern Mediterranean troops stationed on the island during the Roman occupation."
Munley/Manley Surname Project. http://www.familytreedna.com/
"My paternal ancestors were mostly farm workers or self-employed craftsmen of no great social standing. I can trace my line back to the 18th century (confirmed) and as far back as the late 1500s (conjectural). Our Y-DNA Haplogroup of J-M205 (J2b1, old J2b1b) would be considered, quite unambiguously, a potential "Roman Ancestry" DNA signature, being connected in earlier times to the Greek and Thracian Settlements in the Mediterranean basin. As a matter of fact, when confronted with the J2 haplotype by one of his British customers, even Dr. Bryan Sykes of Oxford Ancestors eventually suggested a Roman origin."
K. Pople, Pople Family Association.
"The J2 haplogroup came to England either through middle Eastern Roman soldiers who were stationed on the island (most common explanation), through Sephardic traders (not many of those), or through the migration to the island of gypsies in the 16th century (only being thought of as a possible source very recently)."
Harvey Genealogy. http://history.earthsci.
"As to my family, My Worden branch came from Lancashire England and I can only go back to the late 1400s or early 1500s. The most likely probability is that we are descended from a Roman soldier who married a Saxon woman. There was a Roman retirement villiage right at the area we came from."
mr. Worden, Haplogroup J2b2, 2010.
"As far as our haplogroup goes, J2b2 is still something of a mystery - it's scattered all over Europe, including England, and I have assumed that it had to do with the spread of the Roman Empire. While J (and J1/J2) originated in the middle east and spread in several directions (even to India) it appears that J2b (and J2b2 particularly) seem to be mostly concentrated in Europe, and have probably been there for a long time."
mr. Goodman, Haplogroup J2b2, 2010.
Other Links;
Y-DNA Haplogroup J2 M172, quotes & links. List compiled by R.H.A. Sanders, 2013.
I have made some images and websites which might be of interest to you considering your J2 Blog.
https://www.facebook.com/
http://www.youtube.com/user/
http://www.flickr.com/photos/
http://imageshack.us/user/
https://www.facebook.com/
http://www.youtube.com/user/
http://www.flickr.com/photos/
http://imageshack.us/user/
I hope they prove useful for your blog.
Thanks to Mr. Robert H.A. Sanders for sharing the maps and info above.
Admin & Creator
Admin & Creator
Haplogroup J2 (Y-DNA)
 OriginsHaplogroup J2 is thought to have appeared somewhere in the Middle East towards the end of the last glaciation, between 15,000 and 22,000 years ago. Its present geographic distribution argue in favour of a Neolithic expansion from the Fertile Crescent. This expansion probably correlated with the diffusion of domesticated of cattle and goats (starting c. 8000-9000 BCE) from the Zagros mountains and northern Mesopotamia, rather than with the development of agriculture in the Levant (which seems to have been linked to haplogroup G and perhaps also E1b1b). A second expansion of J2 could have occured with the advent of metallurgy (also from Anatolia and Mesopotamia) and the rise of some of the oldest civilisations. Quite a few ancient Mediterranean and Middle Eastern civilisations flourished in territories where J2 lineages were preponderant. This is the case of the Hattians, the Hurrians, the Etruscans, the Minoans, the Greeks, the Phoenicians (and their Carthaginian offshoot), the Israelites, and to a lower extent also the Romans, the Assyrians and the Persians. All the great seafaring civilisations from the middle Bronze Age to the Iron Age were dominated by J2 men. There is a distinct association of ancient J2 civilisations with bull worship. The oldest evidence of a cult of the bull can be traced back to Neolithic central Anatolia, notably at the sites of Çatalhöyük and Alaca Höyük. Bull depictions are omnipresent in Minoan frescos and ceramics in Crete. Bull-masked terracotta figurines and bull-horned stone altars have been found in Cyprus (dating back as far as the Neolithic, the first presumed expansion of J2 from West Asia). The Hattians, Sumerians, Babylonians, Canaaites, and Carthaginians all had bull deities (in contrast with Indo-European or East Asian religions). The sacred bull of Hinduism, Nandi, present in all temples dedicated to Shiva or Parvati, does not have an Indo-European origin, but can be traced back to Indus Valley civilisation. Minoan Crete, Hittite Anatolia, the Levant, Bactria and the Indus Valley also shared a tradition of bull leaping, the ritual of dodging the charge of a bull. It survives today in the traditional bullfighting of Andalusia in Spain and Provence in France, two regions with a high percentage of J2 lineages. Geographic distributionDistribution of haplogroup J2 in Europe, the Middle East & North Africa 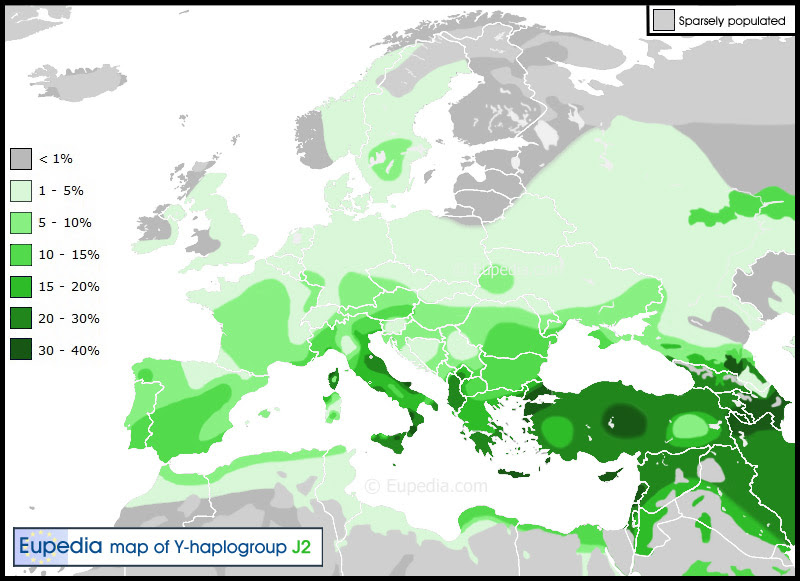 The world's highest frequency of J2 is found among the Ingush (88% of the male lineages) and Chechen (56%) people in the Northeast Caucasus. Both belong to the Nakh ethnic group, who have inhabited that territory since at least 3000 BCE. Their language is distantly related to Dagestanian languages, but not to any other linguistic group. However, Dagestani peoples (Dargins, Lezgins, Avars) belong predominantly to haplogroup J1 (84% among the Dargins) and almost completely lack J2 lineages. Other high incidence of haplogroup J2 are found in many other Caucasian populations, including the Azeri (30%), the Georgians (27%), the Kumyks (25%), and the Armenians (22%). Nevertheless, it is very unlikely that haplogroups J2 originated in the Caucasus because of the low genetic diversity in the region. Most Caucasian people belong to the same J2a4b (M67) subclade. The high local frequencies observed would rather be the result of founder effects, for instance the spread of chieftains and kings' lineages through a long tradition of polygamy, a practice that the Russians have tried to supress since their conquest of the Caucasus in the 19th century. Outside the Caucasus, the highest frequencies of J2 are observed in Cyprus (37%), Crete (34%), northern Iraq (28%), Sicily (26.5%), Lebanon (26%), Turkey (24%, with peaks of 30% in the Marmara region and in central Anatolia), South Italy (23.5%), Bulgaria (20%), Albania (19.5%), and continental Greece (19% excluding northern Greece), as well as among Jewish people (19 to 25%). One fourth of the Vlach people (isolated communities of Romance language speakers in the Balkans) belong to J2, which, combined to the fact that they speak a language descended from Latin, suggests that they could have a greater part of Roman (Italian) ancestry than other ethnic groups in the Balkans. History & SubcladesTwo main subclades divide haplogroup J2: J2a (M410) and J2b (M12, M102, M221, M314). Middle-Eastern and European J2aJ2a's strong presence in Italy is owed to the migration of the Etruscans from the Near East to central and northern Italy, and to the Greek colonisation of southern Italy. The Phoenicians, Jews, Greeks and Romans all contributed to the presence of J2a in Iberia. The particularly strong frequency of J2a and other Near Eastern haplogroups (J1, E1b1b, T) in the south of the Iberian peninsula, suggest that the Phoenicians and the Carthaginians played a more decisive role than other peoples. This makes sense considering that they were the first to arrive, founded the greatest number of cities (including Gadir/Cadiz, Iberia's oldest city), and their settlements match almost exactly the higher frequency zone of southern Analusia. The Romans surely helped spread haplogroup J2 within their borders, judging from the distribution of J2 within Europe (frequency over 5%), which bears an uncanny resemblance to the borders of the Roman Empire. The world's maximum concentrations of J2a is in Crete (32% of the population). The subclade J2a4d (M319) appears to be native to Crete. Indian J2aWithin India, J2a is more common among the upper castes and decreases in frequency with the caste level. This can be explained by the assimilation of local J2a (and R2) people from Central Asia by the R1a Indo-European warriors who descended from modern Russia (Sintashta culture) and established themselves for a few centuries in southern Central Asia, immediately north of the Hindu Kush (including the Oxus civilization) before moving on to conquer the Indian subcontinent. J2a would have reached southern Central Asia with the expansion of Middle Eastern people during the Neolithic and mixed with the local hunter-gatherers belonging chiefly to R2 (and possibly some pre-Indo-European R1a). J2bJ2b has a quite different distribution from J2a. J2b seems to have a stronger association with the Chalcolithic cultures of Southeast Europe, and is particularly common in the Balkans, Central Europe and Italy, which is roughly the extent of the European Copper Age culture. Its maximum frequency is achieved around Albania, Kosovo, Montenegro and Northwest Greece. J2b is also found in the Pontic Steppe, the North Caucasus, Central Asia and in South Asia, particularly in India. Its very low frequency in the Middle East though suggests that, unlike for J2a, it was not spread a progresive and continuous diffusion of the Neolithic lifestyle. For this reason, and because it is generally found among the upper castes of India, it is thought that some J2b lineages might have been part of the Indo-Aryan invasions of South Asia (3,500 years ago) alongside R1a1a. It is conceivable that a minority of J2b, G2a3b1 and R1b1b from the Caucasus region migrated to the Volga-Ural region in the early Bronze Age, spreading with them the Proto-Indo-European language and bronze technology to the Caspian steppe before the expansion of this new culture to Central and South Asia (see R1a history). |
Friday, May 27, 2011
Y Distribution Map
Click the link below to see a larger map and article.
http://www.eupedia.com/europe/european_y-dna_haplogroups.shtml
Thursday, May 26, 2011
J2 originated in northern Mesopotamia
Haplogroup J (Y-DNA)
"J is a Middle Eastern haplogroup, divided into the northern J2 and the southern J1. J2 is by far the most common variety in Europe."
Haplogroup J2
"J2 originated in northern Mesopotamia, and spread westward to Anatolia and southern Europe, and eastward to Persia and India. J2 is related to the Ancient Etruscans, (Minoan) Greeks, southern Anatolians, Phoenicians, Assyrians and Babylonians."
"In Europe, J2 reaches its highest frequency in Greece (especially in Crete, Peloponese and Thrace), southern and central Italy, southern France, and southern Spain. The ancient Greeks and Phoenicians were the main driving forces behind the spread J2 around the western and southern Mediterranean."
"J2 is thought to have arrived in Greece from Anatolia in the early Neolithic, or possibly even earlier. J2b perhaps originated in Greece (or in Anatolia ?), like haplogroup E-V13 (see below) to which it is closely linked. The propagation of J2b and E-V13 (as well as a minority of T) follows the diffusion of agriculture across the Balkans, the Danube basin, and until the north of France to the west, and Moldova to the east. Apart from south-east Europe, J2b is also found all around India, but only at moderate levels in between Europe and India."
"The world's maximum concentrations of J2a is in Crete (32% of the population). The subclade J2a8 appears to be native to Crete. J2a also reaches high frequencies in Anatolia and the southern Caucasus. A likely place of origin is northern Mesopotamia."
"Interestingly, J2a* is found as far as India and is largely confined to the upper castes. The Brahmin (priest) caste is made up almost exclusively of haplogroups R1a1, R2, and J2a (although R1a1 makes up two thirds of the lineages). These 3 haplogroups have Bronze Age coalescence time and are thought to represent the gene flow of the Indo-Aryan invasion of the Indian subcontinent about 3,500 years ago."
Haplogroup J1
"J1 is a typically Semitic haplogroup, making up most of the population of the Arabian peninsula. Its highest density is observed in Yemen (72%), which could be its native place. The Muslim conquest of the Middle East, North Africa, and to a lower extent also to Sicily and southern Spain, spread J1 far beyond Arabia, creating a new Arabic world."
"A considerable part of Jewish people belong to J1 and J2, although J2 is more common. J1 is the Cohen Modal Haplotype, meaning that about three quarters of the people called Cohen, Kohen, or a variant belong to a specific J1 haplotype. In the Hebrew Bible the common ancestor of all Cohens is identified as Aaron, the brother of Moses."
Read the full article:
http://www.eupedia.com/europe/origins_haplogroups_europe.shtml
Sumerian to American
"In spite of the importance of this region, genetic studies on the Sumerians are limited and generally restricted to analysis of classical markers due to Iraq's modern political instability. It has been found that Y-DNA Haplogroup J2 originated in Northern Mesopotamia.[18][19] The Sumerians were a non-Semitic people, and spoke a "language isolate"; a number of linguists believed they could detect a substrate language beneath Sumerian. However, the archaeological record shows clear uninterrupted cultural continuity from the time of the Early Ubaid period (5300 – 4700 BC C-14) settlements in southern Mesopotamia. The Sumerian people who settled here farmed the lands in this region that were made fertile by silt deposited by the Tigrisand the Euphrates rivers."
Click below to read the full article:
Tuesday, May 24, 2011
J2a4h Distribution
Map above and data below are by DDugas, link below:
J2a4h 445=10 Geographic distribution
Most distant ancestor locations for confirmed J2 cases carrying DYS 450=9 and 445=10, indicative of subclade L24, L25. Along with another few markers, these markers 450 and 445 distinguish L25 from most other J2a1's who carry 8 and 12 respectively at DYS 450 and 445.
 | Huntingdonshire, UK |
 | St. Pierre, Oloron-Sainte-Marie, FR |
 | Egliswil, Switzerland |
 | Nieuweschans, Netherlands |
 | Tahmek, Yucatan, Mexico |
 | Brazil |
 | Brazil |
 | Campeche, Mexico |
 | Tenancingo, Toluca, Mexico |
 | Amman, Jordan |
 | Jenin, West Bank |
 | Ourense, Spain |
 | Jordan |
 | Lerna |
 | Lerna |
 | Pakistan |
 | Leek, UK |
 | The Marsh, Shropshire, UK |
 | San Luis Potosi, Mexico |
 | Rumskulla, Sweden |
 | Portugal |
 | Crete |
 | Derio, Pais Vasco, Spain |
 | Sisley, Gujarat, India |
 | Luzzara, Italy |
 | Beirut, Lebanon |
 | Peru |
 | Norfolk, UK |
 | Tulkarm, Palestine |
 | Mostardas, RS, Brazil |
 | Chile |
 | Tarhuna, Libya |
 | Akka, Israel |
 | Mexico |
 | Tiruvalla, India |
 | Doha, Qatar |
 | Oman |
 | Nova Siri, Italy |
 | Gelgaudiskis, Lithuania |
 | Southern Belarus |
 | Minsk, Belarus |
 | Warsaw, Poland |
 | Saukenai, Lithuania |
 | Kamianiec, Belarus |
 | Mlawa, Poland |
 | Vienna, Austria |
 | Cmielow, Poland |
 | Babruysk, Belarus |
 | London, UK |
 | Westland, Netherlands |
 | Sandomierz, Poland |
 | Bialystock, Poland |
 | Zabludow, Poland |
 | Mexico |
 | Bogota, Colombia |
 | Frosinone, Italy |
 | Kottayam, India |
 | Sevilla, Spain |
 | Conturbia-Agrate, Novara, Italy |
 | Alcamo, Sicily |
 | Troina, Sicily |
 | San Luis Potosi, Mexico |
 | Fries, Virginia, USA |
 | Lincoln Co. NC, USA |
 | Calera, Zacatecas, Mexico |
 | New York, USA |
 | Iquitos, Peru |
 | Zacatecas, Mexico |
 | Arequipa, Peru |
 | Cochin, India |
 | Penjamillo, Mexico |
 | Feres, Alexandropoulis, Greece |
 | Ad Dammam, Saudi Arabia |
 | Msaken, Tunisia |
 | Canatlan, Durango, MX |
 | Montefortino, AP, Italy |
 | Dvinsk, Latvia |
 | Torres Vedras, Portugal |
 | Sarikamis, Turkey |
 | Erzurum, Turkey |
 | Erzincan, Turkey |
 | Tunceli, Turkey |
 | Tunceli, Turkey |
 | Bitlis, Turkey |
 | Bitlis, Turkey |
 | Bitlis, Turkey |
 | Bursa, Turkey |
 | Sumnu, Turkey |
 | Bilecik, Turkey |
 | Konya, Turkey |
 | Istanbul, Turkey |
 | Istanbul, Turkey |
 | Istanbul, Turkey |
 | Tokat, Turkey |
 | Kastamonu, Turkey |
 | Bayburt, Turkey |
 | Bayburt, Turkey |
 | Bayburt, Turkey |
 | Diyarbakir, Turkey |
 | Kilis, Turkey |
 | Antakya, Turkey |
 | Mersin, Turkey |
 | Kayseri, Turkey |
 | Yozgat, Turkey |
 | Karabuk, Turkey |
 | India-West |
 | Afghanistan, Hazara |
 | India South |
 | Pakistan-India |
 | Iraq |
 | Halab, Syria (Aleppo) |
 | Taraz, Kazakhstan |
 | Dedacov, Slovakia |
 | Saudi Arabia |
 | India |
 | Florianopolis, Brazil |
 | United Arab Emirates |
 | Hassloch, Germany |
 | Unnamed-Iran |
 | Tetuan, Morocco |
 | United Arab Emirates |
 | Jeddah, Saudi Arabia |
 | Al-Hasa, Saudi Arabia |
 | Kuwait |
 | United Arab Emirates |
 | Mecca, Saudi Arabia |
 | Riyadh, Saudi Arabia |
 | Gonabad, Iran |
 | Chitre, Herrera, Panama |
 | Cocachacra, Peru |
 | Salmas, Iran |
 | Tehran, Iran |
 | Arak, Markazi, Iran |
 | Iran |
 | Lebanon |
 | Mazandaran Province, Iran |
 | Varamin, Iran |
 | Iran |
 | Iran |
 | Khomein, Iran |
 | Sabzevar, Iran |
 | Msaken, Tunisia |
 | Riyadh, Saudi Arabia |
 | Barcelona, Spain |
 | Riyadh, Saudi Arabia |
 | Msaken, Tunisia |
 | Censo, Italy |
 | Gorgan, Iran |
 | Al Hidd, Bahrain |
 | Mosul, Iraq |
 | Iraq |
 | Iraq |
 | Iraq |
 | Iraq |
 | Iraq |
 | Iraq |
 | Iraq |
 | Iraq |
 | Iran |
 | Kuwait City, Kuwait |
 | Samarkand, Uzbekistan |
 | abbotabad, Pakistan |
 | Kahramanmaraş, Süleymanlı, Turkey |
 | Malayer, Iran |
 | Marjaayoun, Lebanon |
Palerermo, IT | |
 | Sivas, Turkey |
 | Yemen |
 | Ahmadabad, Iran |
 | Tabriz, Iran |
 | Elazig, Turkey |
 | Baghdad, Iraq |
 | United Arab Emirates |
 | Qatar |
 | Saudi Arabia-Al Qurashi |
 | Doha, Qatar |
 | Cremona, Italy |
 | Panama |
 | Smyrna |
______________________________

































No comments:
Post a Comment